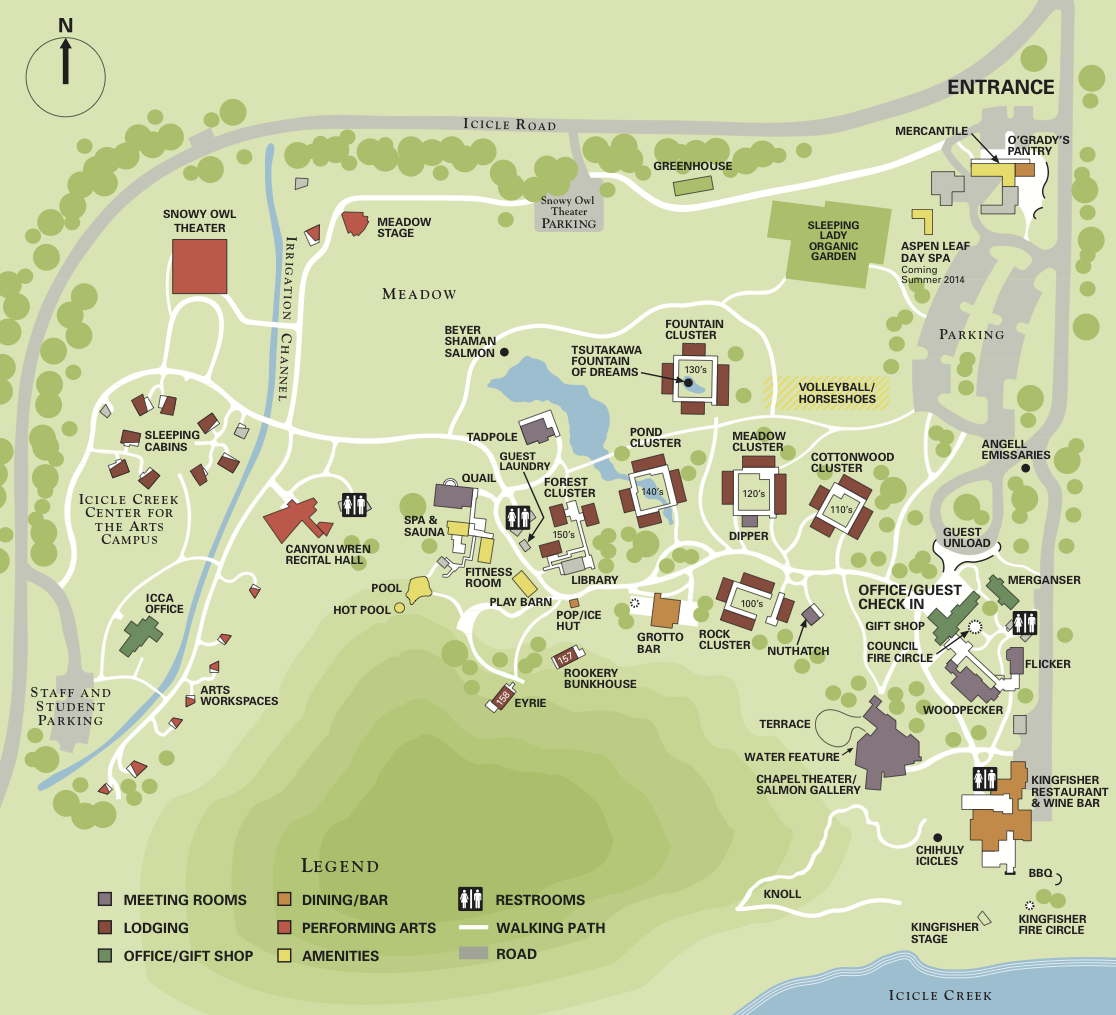
Day 1: Sunday, August 7th
8:45 AM Van Departures from UW Dorms
9:15 AM Van Departures from UW South Center
12:00-2:00 PM Lunch at Kingfisher
- 1:00-1:50 PM Informal Women in Science Meet Up - Find Ora and Rebecca at Kingfisher
- 1:00-1:50 PM Rosetta Commons Executive Board Meeting (Woodpecker)
2:00-3:50 PM Session 1: Chair: John Karanicolas (Chapel)
- 2:00-2:10 PM Chu Wang and Patrick Barth (RosettaCon 2016 Chairs): Introduction and Welcome
- 2:10-2:20 PM Jeffrey Gray (John Hopkins University): RosettaCON Code of Conduct
- 2:20-2:30 PM RosettaCommons Annual Report and Rosetta Software Licensing (Dennis Hanson, University of Washington)
- 2:30-2:50 PM Tanja Kortemme (Kortemme Lab, UCSF): Reshaping Structures and Buildings
- 2:50-3:10 PM Rhiju Das (Das Lab, Stanford University): Vignettes in RNA modeling/design
- 3:10-3:30 PM Ora Furman (Furman Lab, Hebrew University of Jerusalem): Rosetta FlexPepDock going Global
- 3.30-3:50 PM Vladimir Yarov-Yarovoy (Yarov Lab, UC Davis): Design Principls of Membrane Protein Structures
3:50-4:15 PM Coffee Break
4:15-5:10 PM CADRES Session: Chairs: Ora Furman and Xavier Ambroggio (Chapel)
- 4:15-4:30 PM Ora Furman and Xavier Ambroggio (RDG): CADRES assessment
- 4:30-4:40 PM Team 1: Hahnbeom Park, Frank DiMaio, Phil Bradley
- 4:40-4:50 PM Team 2: Aliza Rubenstein, Kristin Blacklock, Hai Nguyen, Sagar Khare, David Case
- 4:50-5:00 PM Team 5: Kyle Barlow, Samuel Thompson, Shane OConnor, Tanja Kortemme
- 5:00-5:10 PM Team 6: Hayretin Yumerefendi, Andrew Leaver-Fay, Brian Kuhlman
5:30-7 PM Dinner at Kingfisher
7:30-8:10 PM Keynote Speaker: David Baker (Chapel): Local encoding of cyclic peptide structures
8:15 PM Poster session 1 (Quail and Tadpole)
Day 2: Monday, August 8th
7:30-9:00 AM Breakfast at Kingfisher
9:00-10:15 AM Session 2: Chair: Vladimir Yarov-Yarovoy (Chapel)
- 9:00-9:35 AM Jie Laing (University of Illinois, Chicago): Computational Transfer Free Energy of Beta Barrel Membrane Protein for Folding and Design
- 9:35-9:55 AM Nik Sgourakis (UC Santa Cruz): Old tricks with new bricks: From protein sample to NMR structure in a matter of days with CHAINS/CS-Rosetta
- 9:55-10:15 AM Tim Whitehead (Michigan State University): Enzyme Design is Unsurprisingly Difficult
10:15-10:40 AM Coffee break
Chapel: Chair: Amanda Duran
- 10:40-11:00 AM Ramesh Jha (Strauss Lab, Los Alamos National Lab): A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor
- 11:00-11:20 AM Gustav Oberdorfer (Baker Lab, UW): Parametric design of helical barrels and pore-like assemblies
- 11:20-11:40 AM Alex Carlin (Siegel Lab, UC Davis): Education and automation integrated into an engineering process for enzyme design
- 11:40-12:00 PM Gideon Lapidoth (Fleishman Lab, Weizmann Institute of Science): Using Evolutionary Principles to Design Novel Enzymes
Woodpecker: Chair: Julia Koehler
- 10:40-11:00 AM Jason Lai (Barth Lab, ): Modeling water-mediated interactions in transmembrane proteins with hybrid solution
- 11:00-11:20 AM Jittasak Khowsathit (Karanicolas Lab, University of Kansas): Computational design of ligand-activated antibodies
- 11:20-11:40 AM Aaron Frank (University of Michigan): Accurate RNA Structure Elucidation Using Chemical Shift-Guided Modeling: Are We There Yet?
- 11:40-12:00 PM Xiang Feng (Barth Lab): Computational design of ligand binding membrane receptors with high selectivity
12:00-2:00 PM Lunch at Kingfisher
2:00-3:00 PM Session 4: Chair: Sagar Khare (Chapel)
- 2:00-2:20 PM Frank DiMaio (University of Washington): Energy function optimization guided by small-molecule and macromolecular data
- 2:20-2:40 PM Sagar Khare (Rutgers University): Design of stimulus-responsive enzymes
- 2:40-3:00 PM Vatsan Raman (University of Wisconsin, Madison): Accerlating design-build-test cycles with design small molecule biosensors
3:00-3:20 PM Coffee Break
3:30-5:10 PM XRM Team Session: Chair: Jens Meiler (Chapel)
- 3:20-3:45 PM Andrew Leaver-Fay (University of North Carolina, Chapel Hill): A Crash Course on XML Schema
- 3:45-4:10 PM Frank Teets & Jared Adolph-Bryfogle: Testing and Tutorials: the 2016 Demos XRW
4:10-4:30 PM Coffee Break
4:30-5:10 PM Session 5: Chair: Jens Meiler (Chapel)
- 4:30-4:50 PM Vikram Mulligan (University of Washington): Designing heterochiral peptides
- 4:50-5:10 PM Rich Bonneau (NYU): Peptidomimetic Design
5:30-7:00 PM Dinner at Kingfisher
- 6:15 PM Meeting for students/PIs attending the Grace Hopper Conference
7:00-7:45 PM Session 6: Chair: Brian Kuhlman (Chapel)
- 7:00-7:05 PM Presentation of Rosetta Service Award
- 7:05-7:45 PM Invited Talk: Bill Degrado (UCSF): De Novo Design, Apllications and Opportunities
7:45 PM Poster session 2 (Quail and Tadpole)
10:50 PM Carcassonne match (Kingfisher Patio)
Day 3: Tuesday, August 9th
7:30-9:00 AM Breakfast at Kingfisher
9:00-10:20 AM Session 7: Chair: Rich Bonneau (Chapel)
- 9:00-9:35 AM Alessandro Senes (University of Wisconsion, Madison): An experimental and computational strategy to study membrane complex assembly
- 9:35-9:55 AM Possu Huang (Baker Lab, UW and Stanford University): Design of chemically synthesized disulfide bonded mini-protein: de novo TIM barrel from parts
- 9:55-10:15 AM Roland Dunbrack (Fox Chase Cancer Center): Structures of protein kinase autophosphorylation events
10:15-10:40 AM Coffee Break
10:20-12:00 PM Open Discussion Topic Groups
- Conformational and Experimentally Guided Protein Modeling (Green Room Chapel)
- Rosetta Tutorial 2016 (Chapel)
- Energy Functions (Dipper)
- Membrane Proteins (Nuthatch)
- Protein Small Molecules (Grotto Pub)
- Peptidomimetrics and Non-canonical Amio Acids (Salmon Gallery Terrace)
- Antobodies Modeling/Designing (Kingfisher Terrace)
- Protein Assemblies and Protein-based Materials (moved next to room 153)
- Protein-Protein Interactions (Palybarn)
12:00-2:00 PM Lunch at Kingfisher
- 1:00 PM Interns, intern mentors, intern PIs, and NSF PO to meet for post-program discussion and evaluation
2:00-2:50 PM Cyrus Session: Chair: Justin Siegel (Chapel)
2:50-3:20 PM Annual RosettaCON photo followed by Coffee Break
3:20-5:00 PM Session 8 (parallel session: Chapel & Woodpecker)
Chapel: Chair: Parisa Hosseinzadeh
- 3:20-3:40 PM Samuel Thompson (Kortemme Lab, UCSF): Pairing high-throughout screening and multi-state design to map the fitness landscape of functional proteins
- 3:40-4:00 PM Andy Watkins (Das Lab, Stanford University): Refining chemically modified RNA structures with ERRASER 2.0
- 4:00-4:20 PM Stephanie Berger (Baker Lab, UW): Computationally designed, high specificity inhibitors probe BCL2 family proteins activity in cancer
- 4:20-4:40 PM Brian Bender (Meiler Lab): Design of Novel Protein-Ligand Interfaces with RosettaLigand
- 4:40-5:00 PM Shao-Qing Zhang (DeGrado Lab, UCSF): De Novo Design of Multi-nuclear Clusters in Helical Bundles
Woodpecker: Chair: Franziska Seeger
- 3:20-3:40 PM Adi Goldenzweig (Fleishman Lab, Weizmann Institute of Science): Designing stable protein variants of recalcitrant proteins in one shot
- 3:40-4:00 PM Amanda Duran (Meiler Lab): Evaluation of methods to predict membrane protein stability
- 4:00-4:20 PM Nick Marze (Gray Lab, JHU): The New RosettaAntibody
- 4:20-4:40 PM Lars Malmstroem (Malstroem Lab, University of Zuerich): Studying protein-protein interactions in complex samples
- 4:40-5:00 PM Robert Kleffner (North Eastern University): FoldIt Standalone
5:00-5:30 PM Annual Swim Across Icicle Creek
5:30-7:00 PM Dinner at Kingfisher
- Women in Science Mentoring Table: Lead by Rebecca, Una, Ora, and Tanja
7:00-7:40 PM Closing Session (Chapel)
8:00 PM PI Meeting (Fire Pit by the Creek)
Day 4: Wednesday, August 10th
7:30-9:00 AM Breakfast at Kingfisher
9:00 AM Bag lunch, checkout, and van/hike departures
The Rosetta Design Group thanks all Participants, the RosettaCommons Community, the University of Washington, and the Sleeping Lady Mountain Retreat for making this event possible. |
||

About the Rosetta Design GroupThe Rosetta Design Group LLC was established in 2007 to bridge the gap between the RosettaCommons academic community and industry. |
|
| Contract R&D in modeling and bioinformatics The Rosetta Design Group provides services ranging from assistance for computational scientists in protocol development to fully out-sourced R&D in structural modeing and bioinformatics for computational and experimental groups whose demands exceed their in-house capacities. |
|
| Rosetta Design Group Website: http://www.rosettadesigngroup.com ROSETTA resources, events, jobs, the Macromolecular Modeling Blog™, and more. |


Industry Participants
 |
 |
 |
Adam Chamberlin Ambry Genetics |
Steven Lewis Cyrus |
Nasos Dousis Moderna Therapeutics |
 |
 |
 |
David Nannemann EMD Serono |
Christopher Meyer |
Zhifeng Kuang AFRL |
 |
 |
 |
Roland Pache Novozymes |
Andrea Halweg-Edwards |
Lei Shi Monsanto |
 |
 |
|
Ryan Ferrao Genentech |
Kyle Roberts Arzeda |


Rosetta Commons Participants
- DiMaio Lab
- Brandon Frenz
- Ryan Pavlovicz
- Frank DiMaio
- Gray Lab
John Hopkins University - Nick Marze
- Jeff Gray
- Sergey Lyskov
- Jeliazko Jeliazkov
- Shourya Sonkar Roy Burman
- Rebecca Alford
- Sofia Bali
- Elizabeth Lagesse
- Michael Pacella
- Jason W. Labonte
- Sammond Lab
National Renewable Energy Laboratory - Deanne Sammond
- Nicholas Sarai
- Das Lab
Stanford CA - Rhiju Das
- Andrew Watkins
- Aaron Frank
- Joseph Yesselman
- Kalli Kappel
- Chu Lab
Peking University - Haobo Wang
- Chu Wang
- Yuan Liu
- Cooper Lab
Northeastern University - Robert Kleffner
- Seth Cooper
- Bonneau Lab
NY University - Richard Bonneau
- Timothy Craven
- P. Douglas Renfrew
- Julia Koehler Leman
- Ryan Butcher
- Leif Halvorsen
- RosettaDesignGroup
- Xavier Ambroggio
- Judith Mueller
- Kuhlman Lab
University of North Carolina - Andrew Leaver-Fay
- Brian Kuhlman
- Mahmud Hussain
- Minnie Langlois
- Stephan Kudlacek
- Jack Maguire
- Sharon Guffy
- Frank Teets
- Ann Cirincione
- Jiang Lab
UCLA - Lin Jiang
- Binsen Li
- Gayatri Nair
- Lindert Lab
Ohio State University - Jacob Bowman
- Sumudu Leelananda
- Steffen Lindert
- Melanie Aprahamian
- Sgourakis Lab
UC Santa Cruz - Nik Sgourakis
- Alison Barrett
- Alastair Fyfe
- Roger Volden
- Fleishman Lab
Weizmann Institute of Science - Ravit Netzer
- Shira Warszawski
- Adi Goldenzweig
- Gideon Lapidoth
- Gront Lab
University of Warsaw - Aleksandra Dawid
- Dominik Gront
- Strauss Lab
Los Alamos National Lab - Charlie Strauss
- Ramesh Jha
- Senes Lab
University of Wisconsin - Alessandro Senes
- Samson Condon
- London Lab
Weizman Institute - Nir London
- Daniel Zaidman
- Max Planck Institute for Biophysical Chemistry
- Colin Smith
- Yarov-Yarovoy Lab
UC Davis - Vladimir Yarov-Yarovoy
- Ruqiang Liang
- Jan Maly
- Ian Kimball
- Phuong Nguyen
- Khare Lab
Rutgers University - Sagar Khare
- William Hansen
- Brahm Yachnin
- Laura Azouz
- Aliza Rubenstein
- Kristin Blacklock
- CoMotion
- Dennis Hanson
- Bradley Lab
Fred Hutchinson Cancer Research Center - Phil Bradley
- Procko Lab
University of Illinois (Urbana-Champlain) - Jihye Park
- Erik Procko
- Meiler Lab
Vanderbilt University - Steven Combs
- Rocco Moretti
- Jens Meiler
- Georg Kuenze
- Alex Sevy
- Amanda Duran
- Brian Bender
- Amandeep Sangha
- Alyssa Lokits
- Marion Sauer
- Corn Lab
UC Berkeley - Jacob Corn
- Nicolas Bray
- Malmstroem Lab
University Zurich - Lars Malmstroem
- Hamed Khakzad
- Kortemme Lab
UC San Fransisco - Tanja Kortemme
- Samuel Thompson
- James Lucas
- Amanda Loshbaugh
- Kyle Barlow
- Xingjie Pan
- Kale Kundert
- Anum Glasgow
- Schief Lab
Scripps Research Institute - Jared Adolf-Bryfogle
- Sebastian Rämisch
- Jenny hu
- Jordan Willis
- Dan Kulp
- Sergey Menis
- Bill Schief
- Torben Schiffner
- Furman Lab
Hebrew University - Ora Schueler-Furman
- Nawsad Alam
- Orly Marcu
- Rachel Prorok
- Yuval Sedan
- Correia Lab
EPFL - Bruno Correia
- Fabian Sesterhenn
- Pablo Gainza
- Dunbrack Lab
Fox Chase Cancer Center - Roland Dunbrack
- Maxim Shapovalov
- Simon Kelow
- Peter Huwe
- Whitehead Lab
Michigan State University - Carolyn Haarmeyer
- Tim Whitehead
- Emily Wrenbeck
- Gu Lab
University of Washington - Liangcai Gu
- Karanicolas Lab
Kensas University - John Karanicolas
- Andrea Bazzoli
- Karen Khar
- Jittasak Khowsathit
- David Johnson
- Jin Niu
- Shipra Malhotra
- Dominic Schenone
- DeGrado Lab
UC San Franscisco - Marco Mravic
- William DeGrado
- Shao-Qing Zhang
- Andre Lab
Lund University - Christoffer Norn
- Ingemar Andre
- King Lab
IPD, University of Washington - Karla-Luise Herpoldt
- Carl Walkey
- Khatib Lab
UMass Dartmouth - Firas Khatib
- Baker Lab
University of Washington - David Baker
- Possu Huang
- Fabio Parmeggiani
- Vikram K. Mulligan
- Sidney Lisanza
- Daniel Silva
- Benjamin Basanta
- Bobby Langan
- Matthew Bick
- Brian Koepnick
- Kathy Wei
- Franziska Seeger
- Chunfu Xu
- Ariel Ben-Sasson
- Zibo Chen
- Cassie Bryan
- Scott Boyken
- Hao Shen
- Isavannah Reyes
- Jiayi Dou
- Parisa Hosseinzadeh
- Alexis Courbet
- Anindya Roy
- Gustav Oberdorfer
- Peilong Lu
- Tamuka Chidyausiku
- Umut Ulge
- Bystroff Lab
Rensselaer Polytechnic Institute - Benjamin Walcott
- Barth Lab
Bylor College of Medicine - Jie Liang
- Xiang Feng
- Jason Lai
- Patrick Barth
- Siegel Lab
UC Davis - Justin Siegel
- Alex Carlin
- Youtian Cui
- Kathryn Guggenheim
- Pamela Denish
- Raman Lab
University of Wisconsin, Madison - Vatsan Raman
- Kyle Nishikawa
- Nick Hoppe
- Megan Leander
- Mills Lab
University of Arizona - Jeremy Mills
- Patrick Gleason
- Patrick Kelly
- Jie Liang Lab
University of Illinois at Chicago - Wei Tian

