A special thanks to our sponsors:
 |
 |
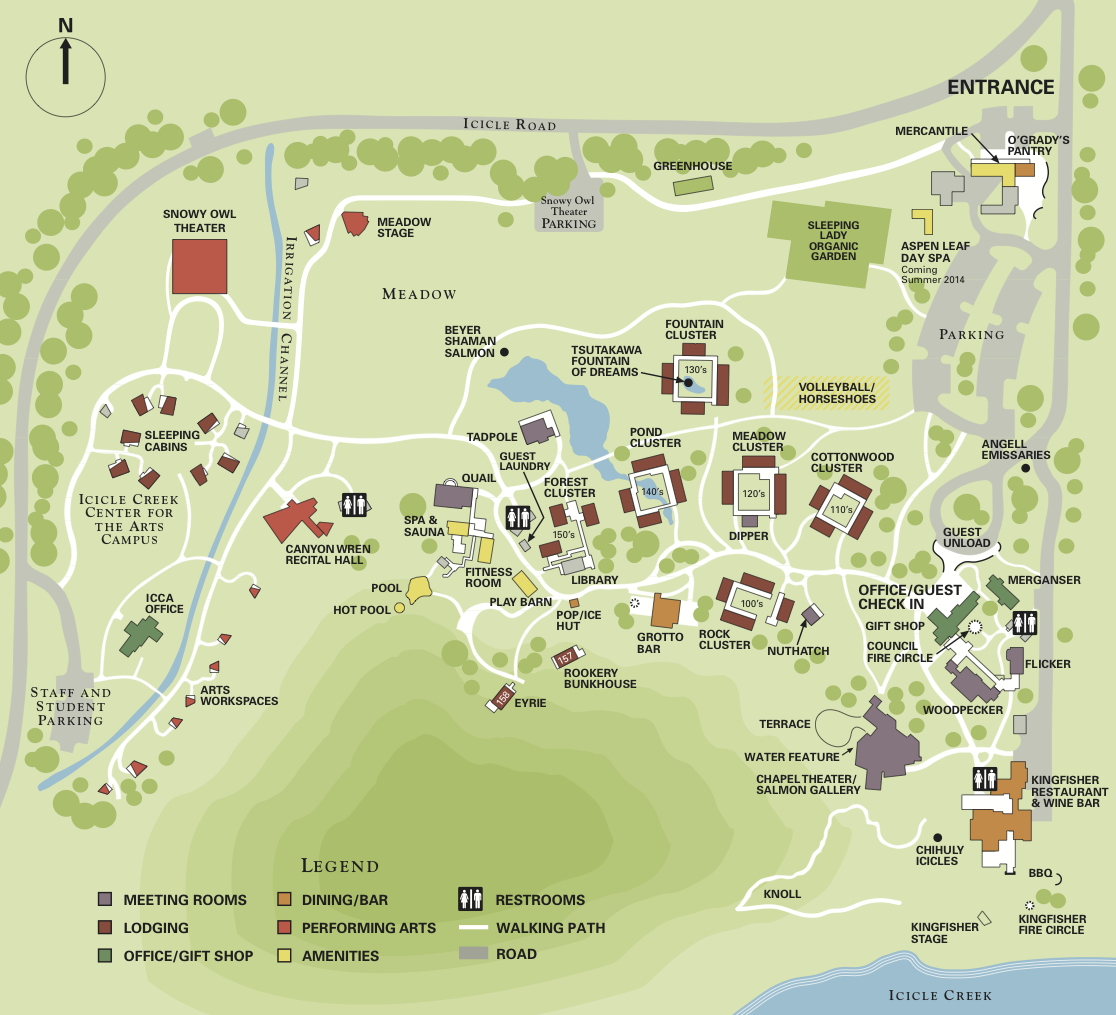
Day 1: Wednesday, July 29th
8:45 AM Van Departures from UW Dorms
9:15 AM Van Departures from UW South Center
12:00-2:00 PM Lunch at Kingfisher
2:00-3:50 PM Session 1: Chair: Frank DiMaio (Chapel)
- 2:00-2:20 PM Introduction and Welcome
- 2:20-2:30 PM RosettaCON Code of Conduct (Karen Khar, Kansas University)
- 2:30-2:40 PM RosettaCommons Annual Report (Dennis Hanson)
- 2:40-3:10 PM RosettaCON Sponsors: Gene Art, Transcriptic, Silicon Mechanics
- 3:10-3:30 PM Jacob Bale - Accurate Design of Multi-component Icosahedral Protein Nanocages -
- 3.30-3:50 PM Amanda Loshbaugh (Kortemme Lab, UCSF) - Engineering Protein Tools That Detect and Respond to Signals in Living Cells -
4:00-5:30 PM Free Time and PI Meeting 1 (Woodpecker)
5:30-7:00 PM Dinner at Kingfisher
- 5:30-7:00 PM Industry Attendee Meet & Greet at Dinner (Kingfisher)
- 5:30-7:00 PM jd3 Discussion at Dinner (at far side of indoor dining area)
7:00-8:00 PM Keynote Speaker: David Baker (Chapel)
8:15 PM Poster session 1 (Quail and Tadpole) Last name beginning with "A" through "k" and people attending the first day only.
9:45 PM Carcassonne match 1 (Kingfisher Patio)
Day 2: Thursday, July 30th
7:30-9:00 AM Breakfast at Kingfisher
9:00-10:20 AM Session 2: Chair: Justin Siegel (Chapel)
- 9:00-10:00 AM Invited Talk 1: Gael McGill -Molecular Visualization: What Does Hollywood Have To Offer?-
10:20-10:40 AM Coffee break
Chapel: Chair: Bruno Correira
- 10:40-11:00 AM Terrence OBrien (Siegel Lab, UC Davis) - A Computational Model of a Terpene Cyclase for Mechanistic Understanding -
- 11:00-11:20 AM Rocco Moretti and Steven Combs (Meiler Lab, Vanderbilt U.) - RosettaDrugDesign - Small Molecule Design Using Rosetta -
- 11:20-11:40 AM Florian Richter (HU Berlin) - A High-activity uAA-tRNA Synthetase from a Rosetta-designed Library -
- 11:40-12:00 PM Joseph Jardine (Schief lab, Scripps) - Development of Germline-Targeting Immunogens -
Woodpecker: Chair: Ingemar Andre
- 10:40-11:00 AM Christopher Bahl (Baker Lab, UW) - De Novo Design of Disulfide-rich Miniproteins: Versatile Scaffolds for High-throughput Engineering -
- 11:00-11:20 AM Aliza Rubenstein (Khare Lab, Rutgers) - A Mean-field Approach to Preducting Multispecificity -
- 11:20-11:40 AM Tina Perica (Kortemme Lab, UCSF) - Multi-state Design and in Vivo Protein Interaction Network Edge Perturbations -
- 11:40-12:00 PM Caitlin Kowalsky (Whitehead Lab, Michigan State University) - Rapid Comprehensive Conformational Epitope Mapping for Antibody-Antigen Structure Prediction -
12:00-2:00 PM Lunch at Kingfisher
- Mentoring Table: Women in Rosetta/Science (Mentors: Tanja Kortemme, Deanne Sammond, and Una Natterman)
2:00-3:00 PM Session 4: Chair: Andrew Leaver-Fay (Chapel)
- 2:00-2:20 PM Lin Jiang (UCLA) - What Can Computational Structural Biology Tell Us About Protein Aggregation in Disease? -
- 2:20-2:40 PM Karen Khar (Karanicolas Lab, Kensas University) - Rescue of Cancer-associated Mutations of p53 Using Rosetta - Best Talk Award Winner
- 2:40-3:00 PM Jorge Fallas (Baker Lab, UW) - Design of De Novo Self-assembling Protein Homooligomers -
3:00-3:20 PM Coffee Break
3:30-5:30 PM Discussion Panel 1: Interfaces for Education, Experts, and Everyone (Chapel)
4:10-4:30 PM Coffee Break
4:30-5:30 PM Session 5: Chair: Brian Kuhlman (Chapel)
- 4:30-4:50 PM Alex Zanghellini (Arzeda) - Enzyme/Strain Design -
- 4:50-5:10 PM Frank DiMaio (UW) - Energy Function Optimization Guided by Experimental Data -
- 5:10-5:30 PM Richard Bonneau (NYU) - Peptidomimetic Design -
5:30-7:00 PM Dinner at Kingfisher
- 6:15 PM Meeting for students/PIs attending the Grace Hopper Conference
7:00-8:00 PM Session 6: Chair: Justin Siegel (Chapel)
- 7:00-7:20 PM Sergey Ovchinnikov (Baker lab, UW) - Using Co-evolution to Predict Unkown Protein Families and Complexes -
- 7:20-8:20 PM Invited Talk 2: Lillian Chong - Molecular Simulation of Protein Binding Pathways with Rigorous Kinetics -
8:20 PM Poster session 2 (Quail and Tadpole) Last name beginning with "L" through "Z" and people attending the second half of the conference only.
9:50 PM Carcassonne match 2 (Kingfisher Patio)
Day 3: Friday, July 31st
7:30-9:00 AM Breakfast at Kingfisher
9:00-10:20 AM Session 7: Chair: Jim Havranek (Chapel)
- 9:00-10:00 AM Invited Talk3: Roberto Chica
- 10:00-10:20 AM Jason Labonte (Gray Lab, JHU) - RosettaCarbohydrates: Expnding the Computational Tools Available to Glycoscientists - Best Talk Award Winner
10:20-12:00 PM Open Discussion Topic Groups
- Conformational and Experimentally Guided Protein Modeling (Green Room Chapel)
- Energy Functions (Dipper)
- Protein Nucleid Acids (Flicker)
- Membrane Proteins (Nuthatch)
- Protein Small Molecules (Grotto Pub)
- Peptidomimetrics and Non-canonical Amio Acids (Salmon Gallery Terrace)
- Antobodies Modeling/Designing (Kingfisher Terrace)
- Protein Assemblies and Protein-based Materials (Qual Porch)
- Protein-Protein Interactions (Palybarn)
12:00-2:00 PM Lunch at Kingfisher
- 1:00 PM Interns, intern mentors, intern PIs, and NSF PO to meet for post-program discussion and evaluation
2:00-2:50 PM Benchmark and Scorefunction Discussion (Chapel)
- 2:00-2:15 PM Sergey Lyskov - Benchmark 2
- 2:15-2:50 PM Discussion Panel 2: History and Future of the Energy Function
2:50-3:20 PM Annual RosettaCON photo and Coffee Break
3:20-4:20 PM Session 8 (parallel session: Chapel & Woodpecker)
Chapel: Chair: Firas Khatib
- 3:20-3:40 PM Enrique Marcos - De Novo Design of Protein Folds with Curved Beta-Sheets Suited for Small-molecule Binding -
- 3:40-4:00 PM Jared Adolf-Bryfogle - A Generalized Framework for Computational Antibody Design -
- 4:00-4:20 PM Charlie E.M. Strauss - Transcription Factors, Library Screens, and Genetically Encoded Materials -
Woodpecker: Chair: Firas Khatib
- 3:20-3:40 PM Cyrus Biotechnology - Enterprise Rosetta on the Cloud -
- 3:40-4:00 PM Brian Kuhlman - Improved detection of Buried Polar Groups -
- 4:00-4:20 PM Christian Schenkelberg - InteractiveROSETTA: A Graphical User-Interface for Rosetta, Prof. Christoph Bystroff Best Talk Award Winner -
4:20-4:40 PM Coffee Break
4:40-5:20 PM Closing Session (Chapel)
- 4:40-5:00 PM Gabriel Rocklin - High Throughput Protein Design at the Edge of Folding Best Talk Award Winner
- 5:00-5:20 PM Awards and Closing Remarks
6:00-8:00 PM Dinner at Kingfisher
8:00 PM PI Meeting 2 (Fire Pit by the Creek) and Rosetta Intro (Chapel)
- Intro to Rosetta Development, or Rosetta is a Cheeseburger (Chapel): Cheeseburger Tutorial, FloppyTail Tutorial, MajorIdeas Tutorial.
10:00 PM Carcassonne match 3 (Kingfisher Patio)
Day 4: Saturday, August 1st
7:30-9:00 AM Breakfast at Kingfisher
9:00 AM Bag lunch, checkout, and van/hike departures
The Rosetta Design Group thanks all Participants, the RosettaCommons Community, the University of Washington, and the Sleeping Lady Mountain Retreat for making this event possible. |
||

About the Rosetta Design GroupThe Rosetta Design Group LLC was established in 2007 to bridge the gap between the RosettaCommons academic community and industry. |
|
| Contract R&D in modeling and bioinformatics The Rosetta Design Group provides services ranging from assistance for computational scientists in protocol development to fully out-sourced R&D in structural modeing and bioinformatics for computational and experimental groups whose demands exceed their in-house capacities. |
|
| Rosetta Design Group Website: http://www.rosettadesigngroup.com ROSETTA resources, events, jobs, the Macromolecular Modeling Blog™, and more. |


Industry Participants
 |
 |
|
Yin He Dorothy Louise Bailey Peter Lee Marie Lee Transcript |
Geoffrey Cassell Thermo Fisher Scientific |
Curran Parker Art Mann Silicon Mechanics |
 |
 |
 |
Aysegul Ozen Genentech |
Benjamin Borgo Agilent |
Greg Lakatos Zymeworks |
 |
 |
 |
Alexandre Zanghellini Arzeda |
Lucas Nivon Javier Castellanos Cyrus |
Shaun Lippow Bayer |
 |
 |
 |
David Nannemann EMD Serono |
Juan Velasquez |
Michael Dolan MSC |
 |
 |
 |
Thomas Holberg Blicher Novozymes |
Krishna Bajjuri |
James Apgar Pfizer |
 |
 |
|
Cara Tracewell Genomatica |
Sally O'Connor |


Academic Participants
- RosettaCommons
- Chong Lab
University of Pittsburgh - Alex DeGrave
- Lillian Chong
- DiMaio Lab
- Brandon Frenz
- Frank DiMaio
- Zibo Chen
- Ryan Pavlovicz
- Gray Lab
John Hopkins University - Brian Weitzner
- Elizabeth Lagesse
- Jason William Labonte
- Jeffrey Gray
- Jeliazko Jeliazkov
- Julia Koehler Leman
- Michael Pacella
- Nick Marze
- Rebecca Alford
- Sergey Lyskov
- Shourya Sonkar Roy Burman
- Sammond Lab
National Renewable Energy Laboratory - Alaksh Choudhury
- Deanne Sammond
- Noah Kastelowitz
- Das Lab
Stanford CA - Caleb Geniesse
- Kalli Kappel
- Meghan Huynh
- Michelle Wu
- Rhiju Das
- Chu Lab
- Yuan Liu
- Cooper Lab
Northeastern University - Seth Cooper
- Bonneau Lab
NY University - Abba Leffler
- Andrew Watkins
- Evan Baugh
- Kamila Kutz
- Kevin Drew
- Leif Halvorsen
- P. Douglas Renfrew
- Richard Bonneau
- Timothy Craveb
- Xiaofei Lin
- RDG
- Xavier Ambroggio
- Kuhlman Lab
University of North Carolina - Andrew Leaver-Fay
- Brian Kuhlman
- Doo Nam Kim
- Frank Teets
- Kevin Houlihan
- Sharon Guffy
- Tim Jacobs
- Jiang Lab
University of Washington - Lin Jiang
- Institute of Chemistry
Humbold University Berlin - Florian Richter
- Sgourakis Lab
UC Santa Cruz - Nik Sgourakis
- Vlad Kumirov
- Khatib Lab
U Mass Dartmouth - Anand Shah
- Firas Khatib
- Mahmood Rashid
- Fleishman Lab
Weizmann Institute of Science - Assaf Elazar
- Gideon Lapidoth
- Liam Longo
- Ravit Netzer
- Sarel Fleishman
- Gront Lab
University of Warsaw - Agata Szczasiuk
- Aleksandra Dawid
- Dominik Gront
- Wieteska
- Strauss Lab
Los Alamos National Lab - Charlie Strauss
- Ramesh Jha
- Yarov-Yarovoy Lab
UC Davis - Drew Tilley
- Ian Kimball
- Phuong Nguyen
- Vladimir Yarov-Yarovoy
- Khare Lab
Rutgers University - Aliza Rubenstein
- Brahm Yachnin
- Kristin Blacklock
- Sagar Khare
- William Hansen
- CoMotion
- Dennis Hanson
- Bradley Lab
Fred Hutchinson Cancer Research Center - Phil Bradley
- Havranek Lab
Washington University (St. Louis) - Adam Joyce
- Chi Zhang
- Jim Havranek
- Wang Lab
Peking University - Chu Wang
- Jinjun Gao
- Procko Lab
University of Illinois (Urbana-Champlain) - Erik Procko
- Jeremiah Heredia
- Invited Speaker
- Gael McGill
- Meiler Lab
Vanderbilt University - Alberto Cisneros
- Alex Sevy
- Amanda Duran
- Amandeep Sangha
- Brittany Allison
- Darwin Fu
- Jens Meiler
- Jessica Finn
- Mingzhao Liu
- Rocco Moretti
- Steven Combs
- Joerg Schaarschmidt
- Corn Lab
- Jacob Corn
- Nicolas Bray
- Malmstroem Lab
ETH Zurich - Lars Malmstroem
- Kortemme Lab
UC San Fransisco - Amanda Loshbaugh
- Eder Davila-Contreras
- Kale Kundert
- Kyle Barlow
- Pradeep Bandaru
- Roland Pache
- Samuel Thompson
- Shane O'Connor
- Tanja Kortemme
- Tina Perica
- Schief Lab
Scripps Research Institute - Alexandra Boukhvalova
- Bill Schief
- Dan Kulp
- Jenny Hu
- Joe Jardine
- Jordan Willis
- Shantanu Bhattacharyya
- Torben Schiffner
- Georgiou Lab
University of Arizona at Tuscon - Oana Lungu
- Furman Lab
Hebrew University - Orly Marcu
- Yuval Sedan
- Correia Lab
UC San Fransisco - Bruno Correia
- Jaume Bonet
- Dunbrack Lab
Fox Chase Cancer Center - Jared Adolf-Bryfogle
- Maxim Shapovalov
- Qifang Xu
- Roland Dunbrack
- Vivek Modi
- Whitehead Lab
Michigan State University - Caitlin Kowalsky
- Emily Wrenbeck
- Justin Klesmith
- Matthew Faber
- Gu Lab
Havard Medical School - Liangcai Gu
- Chica Lab
University of Ottawa - Jamie Davey
- Roberto Chica
- Karanicolas Lab
Kensas University - David Johnson
- Jittasak Khowsathit
- John Karanicolas
- Karen Khar
- Shipra Malhotra
- DeGrado Lab
UC San Franscisco - Thomas Lemmin
- William DeGrado
- Pultz Lab
University of Washington - Clancey Wolf
- Ingrid Swanson Pultz
- Richardson Lab
Duke University - Jane Richardson
- Steven Lewis
- Andre Lab
Lund University - Christoffer Norn
- Ingemar Andre
- Marie Sofie Moeller
- Wojciech Potrzebowski
-
- Baker Lab
University of Washington - Anastassia Vorobieva
- Anindya Roy
- Benjamin Basanta
- Bobby Langan
- Brian Koepnick
- Christopher Bahl
- Chunfu Xu
- Daniel A Silva
- David Baker
- Enrique Marcos
- Franziska Seeger
- Gabriel Rocklin
- Gaurav Bhardwaj
- George Ueda
- Gerard Daniel
- Hahnbeom Park
- Hao Shen
- Hua Bai
- Jason Gilmore
- Jason Klima
- Jiayi Dou
- Jorge Fallas
- Marc Lajoie
- Nathan Rollins
- Peilong Lu
- Per Greisen
- Rachel U. Park
- Ruud van Deursen
- Scott Boyken
- Sergey Ovchinnikov
- Ta-Yi Yu
- Tamuka Chidyausiku
- Tom Linsky
- Una Nattermann
- Vikram K. Mulligan
- Zhizhi Wang
- Bystroff Lab
Rensselaer Polytechnic Institute - Chris Bystroff
- Christian Schenkelberg
- Oluwadamilola Lawal
- Shounak Banerjee
- Barth Lab
Bylor College of Medicine - Melvin Young
- Patrick Barth
- Xiang Feng
- Siegel Lab
UC Davis - Maya Shende
- Steve Bertolani
- Terrence O'Brien
- Wilson Mak
- Morgan Nance

